Cell, Molecular, Health, and Disease
Vector-Borne Diseases
Tamar Carter, PhD, MPH
Vector-borne diseases are a persistent global health threat. A plethora of questions remain about the evolutionary changes that occur within a vector-borne disease system in settings of recent mosquito vector invasions and range expansions. Understanding how mosquito vectors adapt to new environments with different climates, landscapes, and anthropogenic forces can inform models of future spread. Furthermore, understanding the compatibility between invasive vectors and local parasite populations before and after invasions is crucial for predicting the impact of invasions on local malaria epidemiology. Dr. Carter’s research program explores these topics using genomic analysis of natural vector and parasite populations. The lab is currently investigating the phylogeographic history of the malaria mosquito vector Anopheles stephensi’s invasion in east Africa and the environmental factors that facilitate its spread. The lab is also investigating the prevalence and molecular mechanisms of resistance to various classes of insecticides and variation in midgut microbial composition in this species, data that can be leveraged for the development of novel vector control tools. The data science arm of the research program includes the development and/or utilization of genetic and epidemiological databases for modelling geographic variation in malaria transmission intensity. Other projects utilize protein-protein interaction databases to identify the molecular basis of parasite-vector compatibility. The lab also partners with multiple public health and academic institutions in East Africa to plan and conduct molecular surveillance of local vector populations.
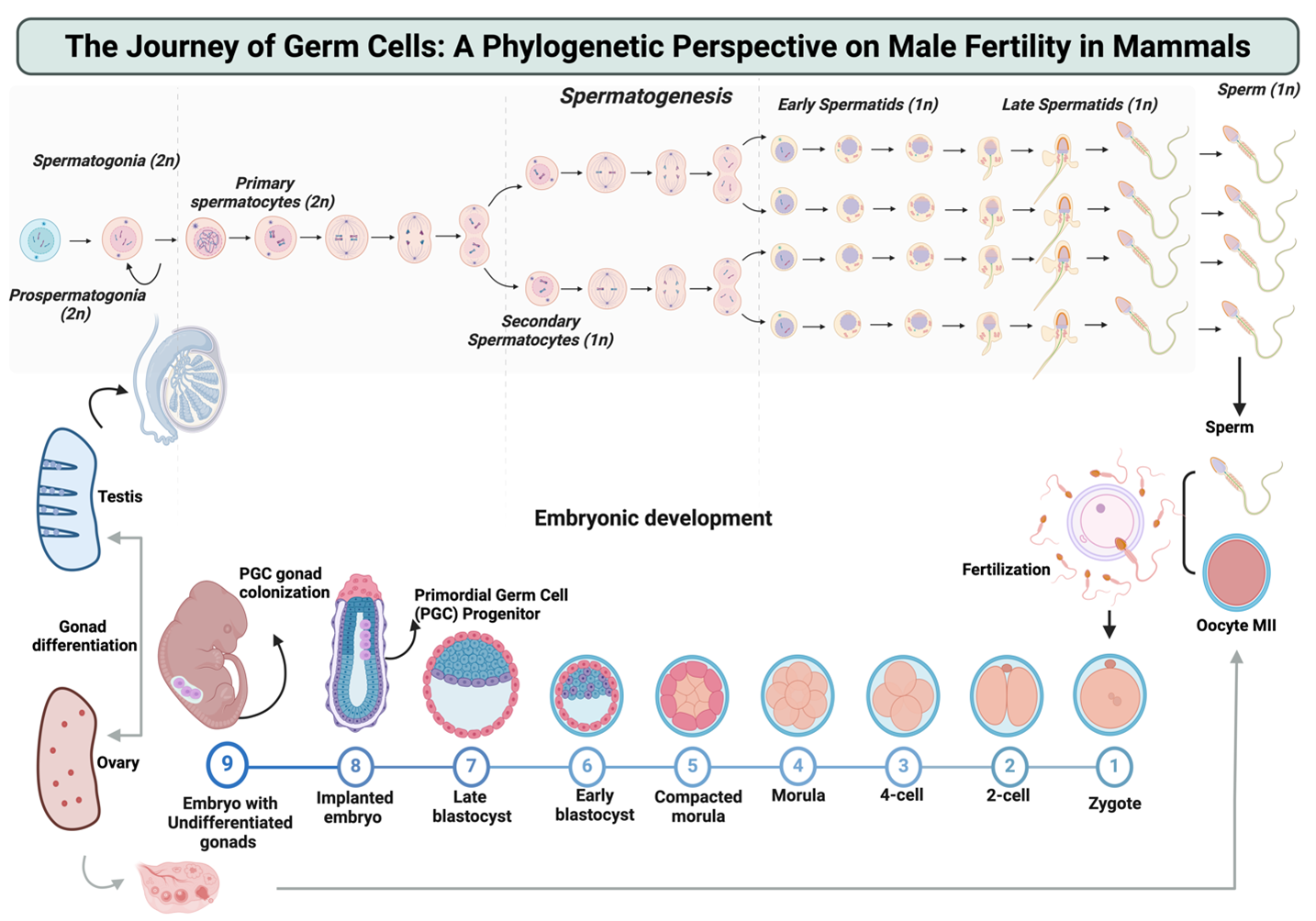
Unlocking the Phylogenetic Pathways of Male Fertility
Mariana Giassetti, PhD, DVM
Our research group is dedicated to investigating male germ cell formation, known as spermatogenesis, and its influence on offspring development in various animal species, including both well-studied models and less-explored non-model organisms. By employing advanced techniques like gene editing and integrative OMICS data analysis, we aim to delve into the intricate molecular processes underlying spermatogenesis in mammals. Our research objectives are threefold:
- Uncover the phylogenetic molecular mechanisms driving male germ cell development.
- Identify factors contributing to male infertility in mammals.
- Reveal the impact of paternal fertility on the early development of animal offspring.
We focus on understanding fundamental biology and translating our findings into real-world applications. Our work holds significance for understanding reproductive processes across species, with implications for biodiversity conservation, invasive species management, and enhancing livestock genetic traits and productivity.
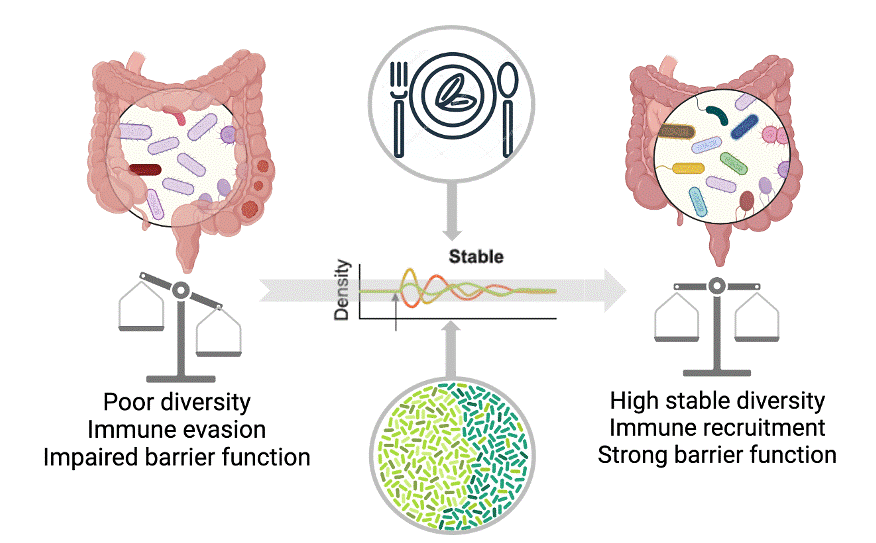
Diet, Microbiome, and Cancer Risk
K. Leigh Greathouse, PhD.
The focus of our research group is understanding the relationship between diet and the microbiome in inflammatory disease, specifically colon cancer. Our goals are to 1) identify the dietary factors that modify the microbiome and their contribution to colon cancer, 2) elucidate the key mechanisms controlling microbiota-host communication, and 3) develop microbial classifiers that improve stratification of patients for colon cancer treatment. Ultimately, our goal is to discover dietary factors and microbial targets for the development of clinical tools to prevent colon cancer development, and reduce morbidity and mortality from colon cancer.
Modulatory Peptides
Christopher Kearney, PhD.
The Kearney Lab studies strategies for the selective control of pathogenic bacteria of the gastrointestinal tract. We are able to kill the pathogen without affecting the commensal bacteria. Using such selective drugs, we are able to avoid the microbial dysbiosis of broad-spectrum antibiotics that leads to adverse health effects. Our "drug" is an antimicrobial peptide with a guide peptide at its N-terminus that binds only to a particular protein on the pathogenic bacterium's surface. Typically, we target virulence factors, such as CagA or VacA of Helicobacter pylori. We deliver the guided antimicrobial peptide (gAMP) by secretion from a probiotic, such as Lactococcus lactis, commonly used in cheese production. This would allow a very advanced peptide drug to be delivered in an inexpensive package, which is fortunate since H. pylori affects low-income populations disproportionately. We also hypothesize that a combination of two or three gAMPs would forestall the development of resistance in the pathogen, similar to the multidrug approach used with antibiotics and anticancer drugs. We also hypothesize that a single gAMP targeting a virulence factor would lead to a pathogen population with mutations in the targeted virulence factor which would attenuate virulence. These hypotheses are presently our main focus.
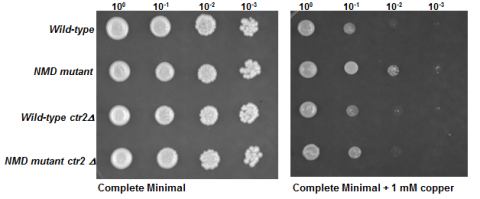
Regulation of Gene Expression
Bessie Kebaara, PhD.
The Kebaara lab is interested in regulation of gene expression at the messenger RNA level (mRNA). We study the recognition and targeting of natural mRNAs by the evolutionarily conserved Nonsense-mediated mRNA decay (NMD) pathway. NMD was first identified as a pathway that degrades premature termination codon (PTC) containing mRNAs therefore preventing the synthesis of truncated proteins. NMD is now also recognized as a pathway that degrades natural mRNAs that primarily encode fully functional proteins. Thus, NMD plays dual roles, one in mRNA surveillance and a second in regulation of gene expression. Our goal is to understand the recognition and targeting of natural mRNAs by the pathway and understand the physiological consequences that result from the degradation of specific natural mRNAs. Currently, we are investigating a subset of mRNAs involved in copper homeostasis.
Cancer Progression and Tissue Homeostasis
Jonathan Kelber, PhD.
There is a strong correlation between solid tumor progression and poor patient survival. Many signaling pathways and molecular/cellular mechanisms that control normal tissue homeostasis are dysregulated in cancer. Our group aims to identify and understand factors that regulate cancer metastasis and therapy resistance, and whether these factors may also control tissue repair.
Biomedical Physiology
Panagiotis Koutakis, PhD.
The Clinical Muscle Biology laboratory investigates the regulation of oxidative stress and mitochondrial function in healthy and clinical populations. We are investigating the effect of different treatments that can improve patients’ health outcomes and provide evidence for new therapeutic treatments. Particularly, we are exploring molecular pathways and cellular properties of ischemia/reperfusion injury in both in-vitro and in-vivo models.
Developmental Cell Biology
Myeongwoo Lee, PhD.
Experimental model system: The nematode Caenorhabditis elegans
Cell-to-extracellular matrix interaction: In many pathological conditions such as metabolic diseases and tumor metastasis, cells often lose their abilities to communicate with their environment. Tissue cells are surrounded by the complex of fibrous proteins and sugars, called the extracellular matrix (ECM). Cells connect to the ECM via a cell surface receptor, integrin. The cell-ECM interaction is mediated by integrin molecules and plays crucial roles in cell organization such as cell adhesion, migration, and differentiation. Our lab studies the function of integrin in vivo. The nematode C. elegans possesses pat-3 β integrin which plays an essential role during embryogenesis. Our studies have shown that pat-3 β integrin is required for the development of gonads, neurons, and the hypodermis. In particular, we are interested in knowing the downstream molecules interacting with integrins in tissue specific ways. Using a gene editing technique, the CRISPR-CAS9 system, we are generating mutations in candidate genes and characterizing genetic or biochemical links to integrin.
Host-pathogen interaction: C. elegans feeds on soil bacteria for reproduction and survival. The research community reports that C. elegans stops reproduction or displays avoidance when it encounters unfavorable conditions such as scarce food or a toxic environment. Our analysis shows that a cell component derived from bacteria appears to stimulate egg laying, a serotonin-dependent reproductive behavior, while it also stimulates the movement of worms. It appears that this bacterial molecule instructs the nematode to lay eggs and move its body. We found that the heterotrimeric G proteins and the Toll-like receptor are involved in recognizing and responding to bacterial cues. Our lab is also interested in learning how such behavioral cues are translated into the stimulation of a particular behavior in the nervous system. To do that, we screen for many mutations in sensory receptors and signaling to identify the genes linked to such behaviors. This study will define molecular mechanism of food or pathogen sensing, which is important for recognizing good or bad conditions for survival.
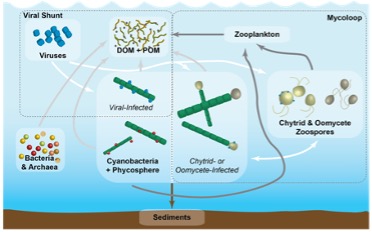
Aquatic Microbial Ecology
Katelyn McKindles, PhD.
The McKindles Lab utilizes a combination of environmental sampling and lab culturing to explore fundamental ecological processes in aquatic microbial systems and applications for harmful algal bloom mitigation and control. Lab members will become familiar with molecular techniques such as PCR, qPCR, RT-PCR, DNA/RNA extraction, metagenomics and metatransciptomics. They will also learn how to sample in the field, generate novel model systems from the environment, and generate and maintain laboratory cultures.
Disease Vector Sensory Biology and Neuroethology
Jason Pitts, PhD.
Arthropod-borne diseases continue to plague developing regions and are being recognized as rising threats to public health in developed nations. Anthropogenic factors such as trade, international travel, global climate change, and habitat modification are major contributors to the spread of vectors and disease-causing agents. Moreover, resistance to pesticides combined with unsustainable control programs has eroded the efficacy of current methods of vector control. New insights into the biology of disease-transmitting insect and tick species are acutely needed in order to combat their devastating effects on human health and economic prosperity.
Our lab investigates the sensory neuronal basis for behaviors in disease-transmitting arthropods, especially mosquito vectors of arboviruses like Dengue and Zika. Of particular interest are the pathways that contribute to chemical- and temperature-oriented behaviors such as host seeking, nectar feeding and oviposition site selection. One of our major goals is to understand complex biological systems by employing a range of techniques including gene expression, neurophysiology, and animal responses to sensory stimuli. One long-term objective of our efforts will be to contribute to reductions in human and animal disease transmission at local, national, and regional levels by improving surveillance and control methods that can be integrated into existing pest management programs.
Vector Biology
Cheolho Sim, PhD.
Currently, the Sim lab focuses on vector biology using a combination of genetic, molecular, cell biological and bioinformatical approaches. The two broad areas of work in our laboratory are (1) molecular regulation of diapause in Culex pipiens and (2) mechanisms of sex determination and sex ratio distortion in Culex pipiens complex species, which are the primary vector for West Nile encephalitis, Eastern equine encephalitis, and many arboviruses, as well as lymphatic filariases.
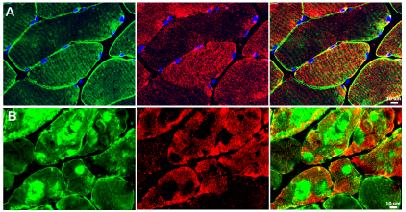
Neurobiology and Cell Signaling
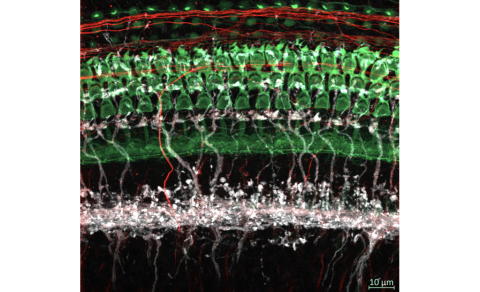
Dwayne Simmons, PhD., Department Chair
The Simmons lab investigates the regulation of calcium signaling during inner ear development, hearing loss, and nerve regeneration using transgenic murine models.
Research interests in the Simmons laboratory concern both developing and aging brain function related to hearing and balance, neurodegeneration, and neuro-immune responses using murine transgenic models. The Simmons laboratory investigates how the regulation of calcium signaling contributes to hearing loss, development and maturation of sensory organs, and peripheral nerve regeneration. Calcium signals can be regulated by specialized protein buffers that bind calcium ions. We have focused our studies on the function of an EF-hand calcium binding protein, oncomodulin (Ocm), which is a member of the parvalbumin gene family. Its distribution is highly restricted, being mostly limited to a subset of sensory hair cells in the mammalian inner ear and elsewhere to a subset of immune cells. Targeted deletion of Ocm in mice leads to an early, progressive hearing loss and to slowed or delayed nerve regeneration. Ocm is expressed in axotomized dorsal root ganglia (DRG) and appears to stimulate axonal outgrowth in DRG neurons. The Simmons laboratory also investigates how specialized sensory cells and their innervation patterns are refined by calcium signals during early stages of development. In the inner ear, calcium signals may be triggered by nicotinic synapses. We are interested in studying these nicotinic synapses in the developing and aged inner ear.
Epigenetics and Cancer Biology
Joseph Taube, PhD.
Outgrowth of disseminated metastases is the major cause of mortality in cancer patients. In the Taube lab, we are investigating the molecular pathways and cellular properties which enable primary tumor cells to metastasize. We focus specifically on epigenetic contributors to cellular plasticity including microRNAs and histone-targeted enzymes.
Microbiome Mechanisms & Biological Consequences
Aaron Wright, PhD.
The Wright group is investigating health-relevant host-microbiome-environment interactions at the level of protein structure and activity. We seek to move the understanding of microbiome activities from largely inference-based to function-based by overcoming the limitations of the current genome-to-phenotype paradigm.
The Wright group performs interdisciplinary research in microbiology, chemical biology, systems biology, and functional -omics to study protein function and protein-small molecule interactions in host-associated microbes and microbiomes, and directly in host organisms. Towards mechanistic understanding of host-microbe-environment interaction mechanisms, the Wright group focuses on: identifying and validating the mechanisms driving microbial functions, interactions and spatial dynamics, and response to perturbation in microbiomes; revealing the biochemical mechanisms for drug/xenobiotic metabolism in mammalian tissues and the gut microbiome, and to understand how these metabolic activities change as a result of mammalian development and chemical exposures; characterizing how irritable bowel disease and related pathologies impact carbohydrate metabolism by the gut microbiome; developing novel chemical biology, systems biology, and ‘omics methodologies to enable discoveries in biology at the molecular scale and with functional resolution.
Vaccine Development and Tropical Medicine
Maria Bottazi, PhD. (Baylor College of Medicine)
Our research laboratories focuses on development of a new generation of drugs, diagnostics and vaccines for neglected tropical diseases (NTDs) and neglected infections of poverty, as well as fundamental and applied research against these diseases.
In the area of NTD vaccine development, Texas Children's Hospital in partnership with the National School of Tropical Medicine has established Texas Children's Hospital Center for Vaccine Development.
Over the past 15 years our research has led the development of new vaccines to combat NTDs. Through this research activity a human hookworm vaccine is in phase 1 clinical trials as is a schistosomiasis vaccine. Vaccines against leishmaniasis, Chagas disease, and other soil-transmitted helminths are in development.
Tropical Medicine and Vaccine Delivery
Peter J. Hotez, MD., PhD. (Baylor College of Medicine)
The Hotez lab leads an international team of scientists working to develop vaccines to combat hookworm infection,schistosomiasis, and other infectious and neglected diseases, including Chagas disease, leishmaniasis, and SARS. Hotez co-founded the Human Hookworm Vaccine Initiative (HHVI) in 1999. He is also heavily involved in vaccine diplomacy, i.e., the opportunity of using vaccines as instruments of foreign policy and to promote global peace, especially among poor countries seeking nuclear weapons technology. Further, the Hotez lab is a global health advocate and policymaker in the area of neglected tropical diseases, with an emphasis on providing impoverished populations access to essential and existing medicines for neglected tropical diseases. His activities have helped to promote awareness of neglected tropical diseases as some of the world's most important health and social issues.
CMHD Faculty with Secondary Appointments in Biology
Mary Lauren Benton, Bioinformatics
The explosion of biological data generated in ever larger populations and across multiple -omic platforms holds great promise in understanding the how DNA sequence is tied to both genome function and disease risk. In addition, increasingly sophisticated experimental techniques requires the development of new computational approaches that can handle the scale of such data. The Benton Lab develops integrative computational approaches to mapping the relationship between gene regulatory sequences, gene expression levels, and disease risk.
Erica Bruce, Toxicology of Human Systems
The overarching focus of research performed in my laboratory is answering a variety of mechanistic toxicological questions relating to insult, injury, and healing of human systems. My area of major interest is in areas relating to hypoxia-induced medical conditions occurring from insults, injury, or exposures to environmental contaminates and pharmaceuticals (i.e., traumatic brain injury (TBI), burns, cancer, chronic wound healing).
Patrick Farmer, Metallobiochemistry/Bioinorganics
The Farmer group studies Cu-based drugs for cancer, NO and H2S metabolism and function in mammalian cells, and metalloenzyme reactivity related to flavonol metabolism. A few representative publications:
“Nitrosyl hydride (HNO) replaces dioxygen in nitroxygenase activity of manganese quercetin dioxygenase” Kumar, M.R.; Zapata, A.; Ramirez, A.J.; Bowen, S.K.; Francisco, W.A.; Farmer, P.J. PNAS USA, 2011, 108, 18926-31.
I use paleontology and archaeology as tools to address research questions firmly grounded in biological theory. Recently, I have published the earliest zooarchaeological evidence for early human hunting and scavenging activities, with implications for the evolution of hominin diets, encephalization, foraging ecology, biogeography, and sociality. My other interests include Paleolithic technology, vertebrate paleontology, and reconstructing hominin paleoenvironments.
Bob Kane, Localized Drug Delivery for Immunomodulation in Transplantation
The Kane research group specializes in the synthesis of bioactive small-molecules, designed for localized delivery, for application in transplantation. Synthetic chemists first, we address transplant immunomodulation by synthesizing active small molecules and their derivatives (prodrugs and bioconjugates). We then evaluate these compounds (and strategies for their delivery) in biochemical, cell-culture, and animal models. Many of the compounds that we synthesize modulate pathways in innate immunity, and bioconjugation chemistry (reactions with soluble proteins or intact live tissue) and substrate-activated prodrugs are utilized to achieve localized and sustained delivery.
Ramon Lavado, Environmental Toxicology
Dr. Lavado is an environmental toxicologist that is working toward investigating biological mechanisms involved in the biotransformation of legacy compounds and advancing in vitro approaches to enable resource-efficient environmental monitoring of the aquatic system. His three main research areas within his overall program are: 1) advancement of in vitro approaches to enable efficient environmental monitoring and a reduction in the use of animals in toxicology; 2) endocrine disruption determination in wildlife associated with environmental pollutants; and 3) drug-metabolizing enzymes and their role in the biotransformation of contaminants of emerging concern and their metabolites in vertebrates, including fish and humans.
The focus of the Law Laboratory is to elucidate mechanisms and identify therapeutic strategies for cancer cachexia, a muscle and adipose-wasting syndrome occurring in advanced cancer patients. We are particularly interested in the studying the pathophysiological role of cardiac dysfunction in pre-clinical cachexia, and how altered metabolism and calcium cycling can contribute to impaired cardiac contractility.
Angie, LeRoy, Health and Behavioral Medicine
Dr. LeRoy is the Director and Principal Investigator of The Baylor “HEAL” Lab, Studying HEalth Across the Lifespan. Dr. LeRoy includes a variety of health-related measures in her research studies including biomarkers of immunity, parasympathetic nervous system activity, observational wound healing, and patient-reported outcomes. One of the HEAL lab’s core initiatives is to elucidate the various physiological and neurobiological systems involved in human responses to social separation—particularly in cases when an individual is separated from someone with whom they have formed an attachment relationship (e.g., mother-child dyads, romantic partners), whether it be brief (e.g., temporary physical separation due to travel or quarantine) or prolonged (e.g., following divorce from a partner, or the death of a partner). In addition, the HEAL lab develops and implements interventions to help people heal after social loss.
Jung Hyun Min, Structural Biochemistry and Biophysics of Nucleotide Excision Repair
We seek to understand how cellular DNA repair works by investigating the structures and dynamics of protein-DNA complexes involved in DNA damage sensing and repair using X-ray crystallography and various biochemical/biophysical techniques.
Michael Muehlenbein, Evolutionary Medicine
We are a group of biological and evolutionary anthropologists interested in a variety of research subjects, including evolutionary endocrinology, ecological immunology, reproductive ecology, human life history evolution, behavioral endocrinology, animal behavior and ecology, evolutionary psychology, infectious disease ecology, and emerging infectious diseases.
Christie Sayes, Environmental Health and Material Science
Dr. Christie M. Sayes is a practicing research scientist and consultant in the fields of toxicology, chemistry, material science, and environmental health. Sayes is a subject matter expert in advanced materials, exposure science, health effects, and risk. Her activities include working with partners, collaborators, and trainees in designing studies related to safety-by-design considerations of engineered materials and emerging contaminants used in pharmaceutical, agricultural, and consumer products. Sayes is also interested in occupational safety and environmental transformations of particle systems in complex matrices. She possesses a working knowledge of laboratory science and U.S. regulatory climates. Routine activities include validating alternatives to animal models, zebrafish and rat in vivo models, biological and chemical molecular mechanistic analyses, mass spectrometry, electron microscopy, and statistics. Data sets are always related back to the published literature, compared against appropriate controls, and verified using orthogonal methods.